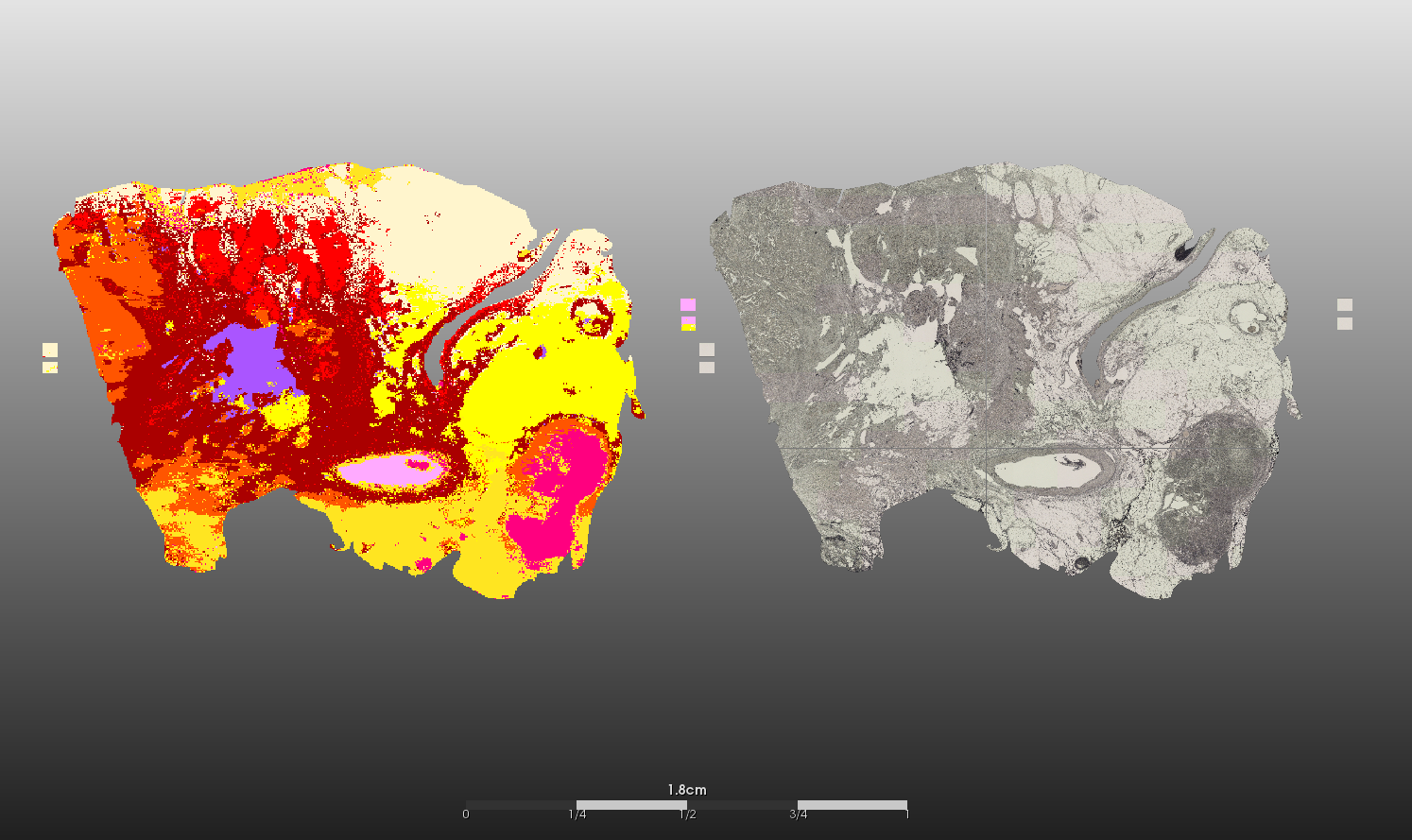
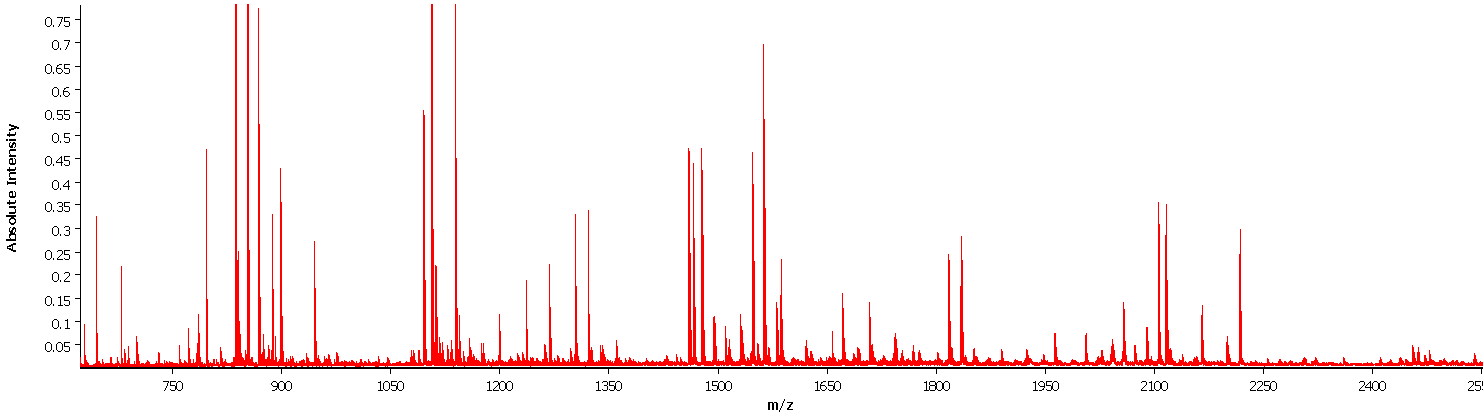
Matrix-associated laser desorption/ionization imaging (MALDI imaging) combines two major fields of bioanalytical methods: molecule analysis using time-of-flight mass spectrometry and the macroscopic anatomy of the tissue compartments. Regions of interest (ROI) of human cancer tissue slides defined by pathologists are translated into mass spectra patterns and can be used for subsequent biostatistical analyses of protein, peptide or lipid composition and quantification. In addition to the knowledge gained from differing mass spectra in e.g. tumor versus normal tissue areas, morphologically indistinguishable tumors can be subdivided and analyzed for molecule-dependent heterogeneity through segmentation of mass intensities at a very high resolution (down to 5x5 micrometers).
In addition to the significance of conventional MALDI analysis for routine diagnostics and for medical research projects using experimental setups, the MALDI Imaging Unit researches bioinformatic workflows and deep learning pipelines to utilize big data e.g. for automation purposes.