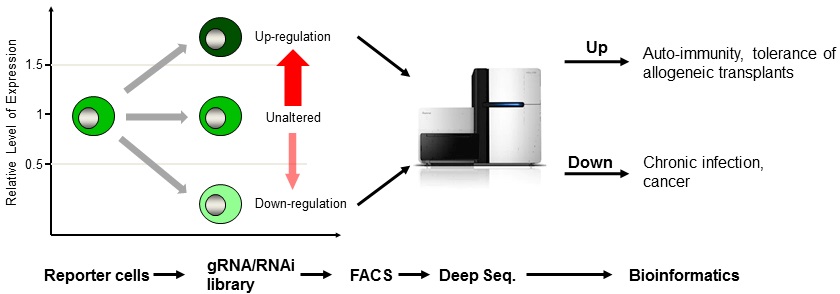
Advances in cancer biology and immunology have paved the way to treating cancer by directing the immune system to target neoplastic cells. This new field of cancer immunotherapy has shown great promise as a treatment strategy in the clinic. As different routes were pursued to direct the immune system to fight malignancies, therapeutic targeting of cancer immune checkpoint molecules proved to be remarkably effective as it diminished cancer immune resistance.
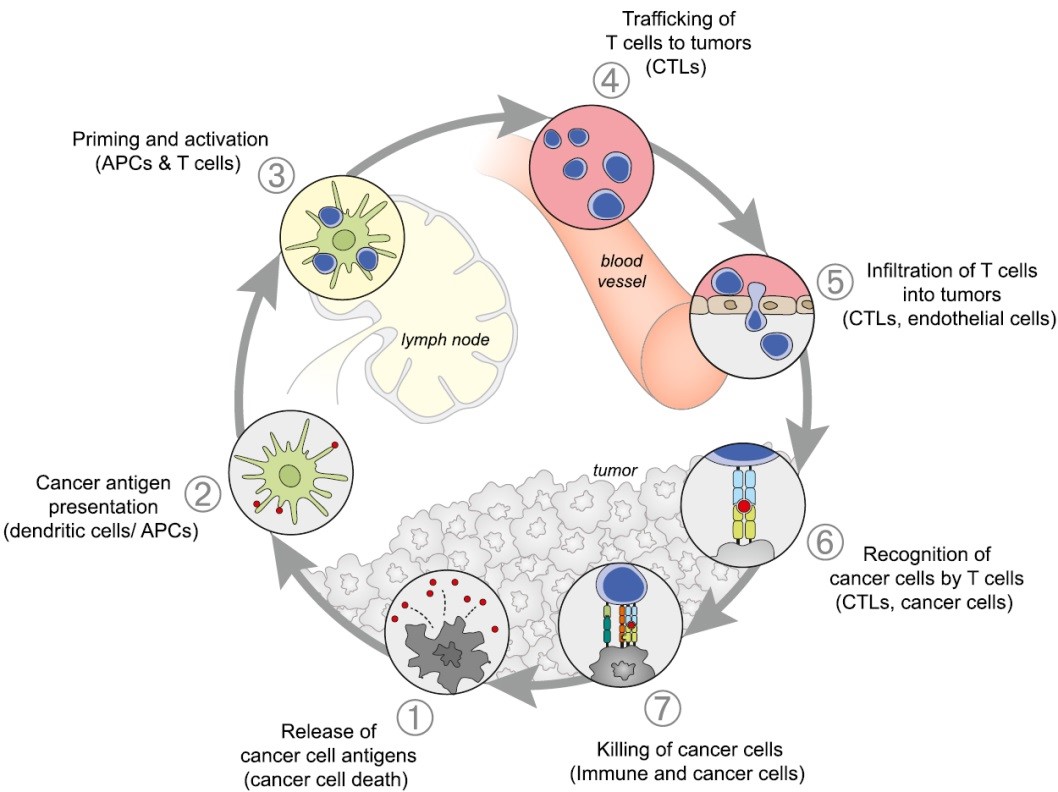
The majority of these therapies benefit from the fact that many tumors are immunogenic and are infiltrated by cytotoxic T lymphocytes (CTLs) expressing T-cell receptors (TCRs) that bind to tumor antigens (Fig. 1). To maintain immune resistance, tumor cells have to develop strategies to inhibit these CTLs. This process involves many interacting partners on both the antigen-presenting cells (APCs) and the T-lymphocytes. Although some key players have been identified such as the protein PD-L1 (programmed cell death ligand 1) most molecular components and their cellular regulation remain elusive.