Computer Assisted Navigation
Computer Assisted Navigation (CAN) in tumor surgical treatment is a rapidly developing research field. The CAN systems enable surgeons to accurately identify and remove tumors while minimizing damage to surrounding healthy tissue to improve oncological and functional outcomes. Our research in this field focuses on improving the speed, accuracy, and precision of soft-tissue navigation systems, as well as developing new methods of generating real and synthetic datasets necessary for training and integrating the obtained ground truth data into not only the intraoperative but also the surgical planning processes. We focus on a wide spectrum of research questions in the field, from machine learning aspects that can improve surgical systems to aspects of clinical intraoperative applications, all in the pursuit of our goal of better-personalized patient outcomes and a higher quality of life for those affected by cancer.
We are researching methods, developing algorithms, and creating datasets for:
- 3D Reconstruction, Depth Estimation, Camera Calibration
- Intraoperative Segmentation
- Rigid and Non-Rigid Registration
- Soft-Tissue Modeling
- User-Interaction & User-Intervention
- Augmented Reality & User-Interfaces
Research Topics
The purpose of intraoperative navigation is to provide the surgeon with real-time information about the location of critical structures, such as blood vessels and nerves, as well as the tumor itself. This information is based on preoperative planning data, such as CT or MRI scans of the patient's anatomy, and is overlaid with the surgeon's view of the surgical field using augmented reality (AR) technology. In soft-tissue navigation, for example in the case of liver surgery, the deformation of organs and tissues can be a significant challenge. This is due to the fact that the organs can shift and change shape during the procedure, making it difficult to accurately track their position in real time. Additionally, different patient poses and breathing patterns can also affect the accuracy of the navigation system.
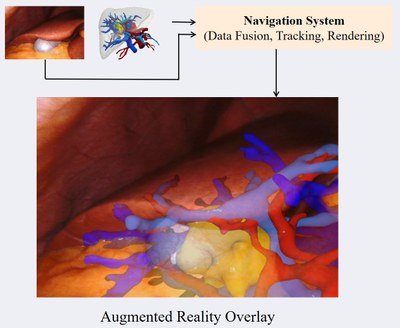
To address these challenges, we are developing new methods that can account for the deformation of organs and tissues. This includes using advanced real-time imaging techniques to provide up-to-date information on the location of critical structures during the procedure. A core focus of our group is using novel machine learning algorithms to predict the deformation of the tissues based on the observations of the procedure, allowing the navigation system to more accurately track their position in real time. We explore the use of deep neural networks to perform registration between the preoperative and intraoperative states. These networks can be trained on a mixture of real and synthetic data, allowing them to learn to find relevant matching features and estimate the deformation between the two states.
Obtaining the data necessary for training and evaluation of algorithms is a major bottleneck in the field of Computer Assisted Interventions (CAI). While other fields mainly struggle with labeling data, CAI has the additional challenges of data privacy, the fact that many cases are outliers, lack of standardization, and lack of ground truth. To illustrate, a ground truth depth map of the intraoperative video would be useful for navigation algorithms, but it is extremely difficult to obtain from surgeries. The main problem in the time-consuming labeling of data is that annotations usually have to be performed by experts.
With these problems in mind, the simulation and generation of synthetic datasets have many advantages. Synthetic data is controllable, (virtually) infinite, accessible, and can easily be shared. Moreover, synthetic data can be generated with a high degree of standardization as well as with a ground truth that is known which can be useful for comparing results between different research groups and for evaluating the accuracy of algorithms.
In our group, various projects utilize synthetic data for training or evaluating machine learning algorithms that operate on real data. We often publish the simulation methods as well as the generated datasets.
Data-driven Simulation Pipelines
There are almost no public datasets available that show the liver of a single patient in multiple deformed states. To drive our machine learning algorithms for navigation and registration, we are developing simulation pipelines to generate synthetic deformations of both real and non-real organs. We build upon physically accurate biomechanical models to generate a vast amount of semi-realistic data which is then used to train fast and powerful neural networks. Simulation pipelines to generate synthetic deformations of the liver and other organs involve complex computational modeling techniques to simulate the behavior in response to different types of deformations. One of the biggest challenges is to construct and refine the physical and deformation models, including information about the organ's geometry, biomechanics, and tissue properties that can accurately capture the complex behavior of these organs.
The goal of our research is to develop simulation pipelines that can generate realistic and diverse synthetic data while also ensuring that the data is labeled correctly and is representative of real operation scenarios. To validate the model and generate deformation ground truth data, we combine the mathematical and computational modeling techniques based on data from real patients, such as medical imagines and physiological data. Further data augmentation techniques, such as random transformations and perturbations and more complex modeling techniques are being combined in our group to generate highly realistic synthetic data.

Neural Rendering
In the surgical domain, obtaining ground-truth data for computer vision tasks is almost always a major bottleneck. This is especially the case for ground truth which would require additional sensors (e.g. depth, camera poses, 3D information) or fine-grained annotation (e.g. semantic segmentation). To this end, we aim at rendering photo-realistic image and video data from simple surgical simulations (e.g. laparoscopic 3D scenes). We utilize and develop methods from emergent areas such as neural rendering and generative image synthesis. The resulting synthetic visual data, along with rich ground truth, can then be used as training or evaluation data for various computer vision tasks (e.g. SLAM, depth estimation, semantic segmentation).

Open Access Datasets
Group Members
- Micha Pfeiffer (Postdoc, Group Leader)
- Maxime Fleury (Scientific Software Development)
- Gregor Just (Scientific Software Development)
- Dominik Rivoir (Doctoral Student)
- Peng Liu (Doctoral Student)
- Reuben Docea (Doctoral Student)
- Johannes Bender (Doctoral Student)
- Bianca Guettner (Doctoral Student)
- Danush Kumar-Venkatesh (Doctoral Student)
- Jinjing Xu (Doctoral Student)
- Stefanie Speidel (Professor, PI)