NCT/UCC MALDI Bildgebungs Unit

Neben der konventionellen MALDI-Analyse für die Routinediagnostik und neuen medizinische Fragestellungen erforscht die MALDI Imaging Unit bioinformatische Workflows und KI-basierte Rechenwege, um diese komplexen Datensätze schnell zu visualisieren.
Forschungsschwerpunkte
- Tumorklassifizierung seltener Tumorarten
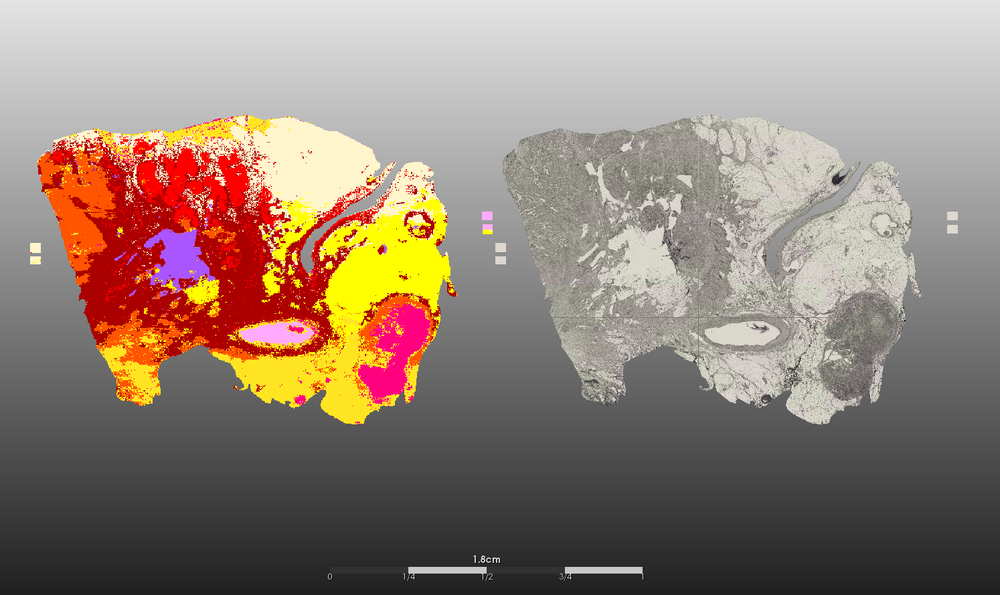
- Segmentierung der (intra-)tumoralen Heterogenität
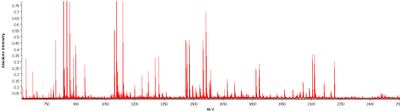
- Identifizierung von Lipid-, Protein- und Peptidmassenspektren neuartiger Tumorsubtypen
- Aufbau einer ständig wachsenden Datenbank mit Massenspektrometriedaten von Tumorgewebe im Vergleich zu normalem Gewebe
- MALDI-Segmentierungspipelines auf Basis von Deep Learning
- 3D-MALDI-Bildgebungsverfahren
Probenarten für die MALDI Bildgebung
- Frisch gefrorenes Gewebe
- Formalin-fixiertes, in Paraffin eingebettetes (FFPE) Gewebe
- Proben mit und ohne enzymatische Verdauung auf Objektträgern
- Eingebettete Organoide, Sphären oder Zellkulturblöcke
Technische Informationen
- Positiver und negativer Flugmodus, erfasst durch linearen oder Reflektordetektor
- Massenbereich (positiver Modus, z. B. Peptide: 600–3200 m/z, Proteine 1000–500.000 m/z)
- Scanbereich: 5 x 5 µm bis 150 x 150 µm resultierende Feldgröße
- Option für automatisierten Batch-Betrieb
- Externe Kalibrierung und Qualitätskontrollen werden bereitgestellt
- Big-Data-Speicher
Koordination
Dr. rer. med. Pia Hönscheid
Gruppenleiterin
E-Mail: pia.hoenscheid(at)ukdd.de
Telefon: +49 (0)351 458 13038
Direktor
Prof. Dr. med. Gustavo Baretton
Direktor Institut für Pathologie
E-Mail: Gustavo.Baretton(at)ukdd.de
Telefon: +49 (0)351 458 3000
Team
Christian Sperling
Medizinisch technischer Assistent
E-Mail: Christian.Sperling(at)ukdd.de
Telefon: +49 (0)351 458 13009
Medizinstudent
E-Mail: maxime.lefloch(at)medforum-dresden.de
Telefon: +49 (0)351 458 3009
Maximilian Weiss
Arzt in Weiterbildung
E-Mail: Maximilian.Weiss(at)ukdd.de
Telefon: +49 (0)351 458 5266